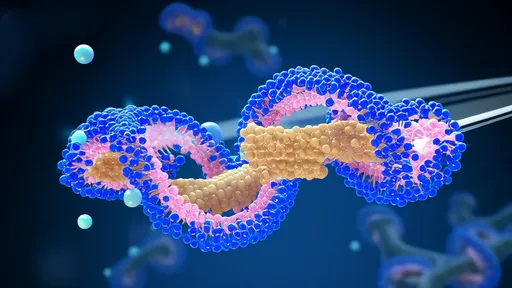
In a groundbreaking study that reshapes our understanding of genome architecture, researchers have uncovered how cohesin dynamically maintains the three-dimensional structure of DNA through a mechanism dubbed the "topological lock." This discovery provides unprecedented insights into how chromosomes organize themselves within the nucleus and how this organization influences crucial cellular processes such as gene regulation, DNA replication, and repair.
The cohesin complex, long known for its role in sister chromatid cohesion during cell division, has now been revealed as a master regulator of chromosomal topology. Using advanced imaging techniques and single-molecule analyses, scientists observed that cohesin acts like a molecular clamp, creating and maintaining loops in DNA by forming a topological embrace around the chromatin fiber. This embrace, or "lock," is far more dynamic than previously thought, with cohesin constantly releasing and re-engaging with DNA in response to cellular signals.
What makes this mechanism particularly remarkable is its precision and adaptability. The topological lock isn't a permanent fixture but rather a fluid system that allows chromosomes to reorganize themselves as needed. When genes need to be activated or silenced, when DNA damage requires repair, or when chromosomes prepare for division, cohesin adjusts its grip, enabling the genome to fold and unfold in a highly regulated manner. This dynamic quality explains how cells can maintain genome stability while allowing the flexibility required for diverse genomic functions.
Researchers employed an innovative combination of CRISPR-based genomic editing, super-resolution microscopy, and in vitro reconstitution experiments to dissect this mechanism. They found that cohesin doesn't simply bind to specific DNA sequences but instead slides along the chromatin, extruding loops until it encounters certain boundary elements. This process, termed "loop extrusion," is powered by ATP hydrolysis and is carefully controlled by a suite of regulatory factors including NIPBL, Wapl, and PDS5 proteins.
The implications of these findings extend across multiple fields of biology. In developmental biology, the topological lock mechanism helps explain how cells maintain their identity during differentiation. In cancer research, it provides new clues about how chromosomal translocations and other aberrations might occur when cohesin regulation goes awry. Furthermore, this discovery offers fresh perspectives on a range of genetic disorders known as cohesinopathies, which include Cornelia de Lange syndrome and Roberts syndrome.
One of the most surprising aspects of the study was the revelation that cohesin's topological embrace can persist through multiple rounds of DNA replication. This challenges previous assumptions that the complex's hold on DNA would be disrupted during S phase. Instead, it appears that cohesin can maintain its grip even as the replication fork progresses, providing continuous organization of the newly synthesized chromatin.
As the scientific community digests these findings, attention is turning to the many remaining questions. How exactly do various cellular signals modulate cohesin's activity? What determines where loops form and how large they grow? And how does this mechanism interact with other elements of nuclear architecture, such as lamins and nuclear pore complexes? Future research directions will likely explore these questions while also investigating potential therapeutic applications of manipulating the topological lock in disease contexts.
This discovery represents a significant leap forward in our comprehension of genome organization. The topological lock model not only explains long-standing observations about chromosome behavior but also provides a unifying framework for understanding how diverse genomic processes are spatially coordinated. As research continues, we can expect this mechanism to reveal even more about the exquisite complexity of life at the molecular level.

By /Aug 7, 2025
By /Aug 7, 2025

By /Aug 7, 2025

By /Aug 7, 2025

By /Aug 7, 2025
By /Aug 18, 2025

By /Aug 7, 2025

By /Aug 7, 2025

By /Aug 7, 2025
By /Aug 7, 2025
By /Aug 18, 2025
By /Aug 18, 2025

By /Aug 7, 2025

By /Aug 18, 2025
By /Aug 7, 2025
By /Aug 18, 2025

By /Aug 7, 2025

By /Aug 7, 2025

By /Aug 7, 2025

By /Aug 7, 2025